Enzyme discovery
Begin your enzyme journey with us, knowing you'll have the expertise and support to see it through to successful commercialization!
The first thing we need is a good starting point.
Sequence overload!
There is a vast expanse of available sequences—spanning billions.
Rest assured, we stand out as exceptional hunters in the realm of enzyme discovery. Our team excels in navigating this complex landscape, employing cutting-edge biodiversity computational tools to efficiently sift through the massive number of sequences.
We recognize that sometimes natural sequences are desired. In such cases, extensive testing of numerous natural candidates is essential to pinpoint those with the desired properties.
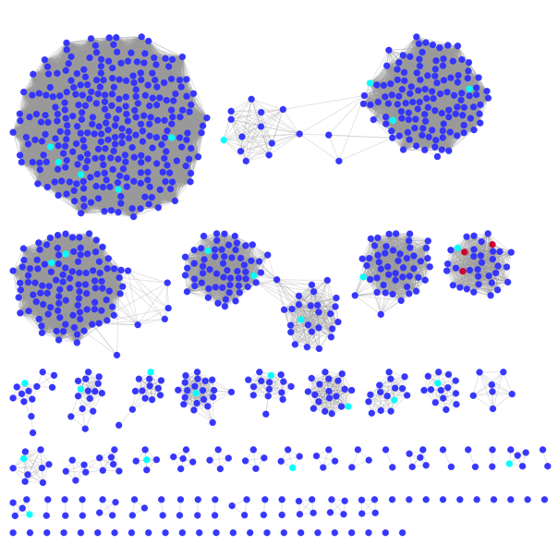
Pictured above: One of our search strategies (divide and conquer) where we cluster the sequence space using bioinformatics algorithms and then explore within each cluster (cyan dots), eventually honing in on the best enzymes for our partners (red dots).
Our approach begins with a clear vision of the end goal. We leverage specialized algorithms designed to predict enzyme activity from sequence and structure, such as optimal temperature and pH ranges. Our tools also include advanced algorithms for identifying enzymes with properties similar to patented ones, while optimizing for freedom-to-operate (FTO). This ensures that we find proteins with comparable performance that are free from existing patents. Additionally, our algorithms can assess potential allergens, toxins, and virulence factors, ensuring that the enzymes we discover are safe and suitable for various applications.
Too few sequences?
A recent addition to our toolkit is generative AI to create novel sequences, opening new avenues for enzyme development.
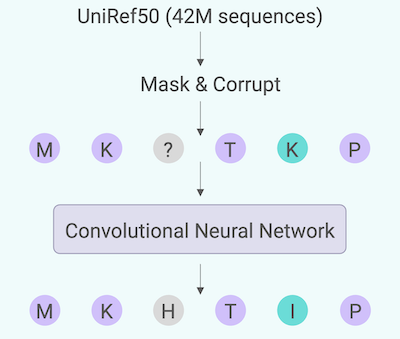
In our testing, these algorithms need only a few sequences to be primed with since they were trained on all of UniProt; however, use of generative AI to expand the sequence space while retaining functionality is still an emerging capability for the field!